Membranes
 (image source)
(image source)
The modeling of biological membranes has been a high priority of Life123 since its inception... and their early implementation
began with the Beta 12 release. The recent Beta 13 rolled out a full implementation of the data structure in 1D,
designed to be biologically relevant and extendable to 2D and 3D, plus functions for data management, visualization and reactions.
Diffusion across membranes (passive transport) is expected to be implemented in Beta 14.
Many of the existing functions to read/modify bin concentrations, have been extended to make allowance for the possible presence of membranes in some bins.
Early visualization functions are included in Beta 13... while full plotly visualization is expected to start in Beta 14.
One guiding criterion to model membranes is the great disparity between the puny thickness of biological membranes (around 4-5 nm)
vs. the overwhelmingly larger size of cells (diameters around 0.1 to 5 µm for prokaryotes and some 10-100 µm for eukaryotes.)
We're talking about ratios (of membrane thickness to cell diameter) of some 1:20 to 1:25,000!
A handy source of typical biological values can be found on this site.
The initial modeling of membranes in Life123 will tentatively be as follows:
- "Membranes", as a term used in Life123, refers to the entire lipid bilayer, and any molecules embedded in it
- In keeping with Life123's guiding principles, no attempt will be made to attain detailed modeling of the biophysics of the membranes; rather, the goal is for a light/relatively minimalist model that still gives adequate guidance on the evolution of the dynamical system (diffusion, reactions, mechanical effects, etc.)
- Membranes are much thinner than bins. In the (rather) unlikely event that the spacial resolution of the bins is so fine as being thinner than membranes, spacial resolution is meant to be adjustable in Life123 – and it will be made large enough around membranes
- At most 1 membrane per bin. In the event of 2 membranes in close proximity, the spacial resolution of the bins can simply be improved, as to keep them in separate bins.
- If multiple membranes find themselves in the same bin, they are consired to be fused.
- Every chemical species will have a special "diffusion rate" (absorption and desorption thru membranes), to model passive transport – and, for now, the same for all membranes
- Active transport by means of reactions with trans-membrane molecules will be modeled in later iterations, probably starting with the modeling of macro-molecules (with multiple binding sites) around Beta 15.
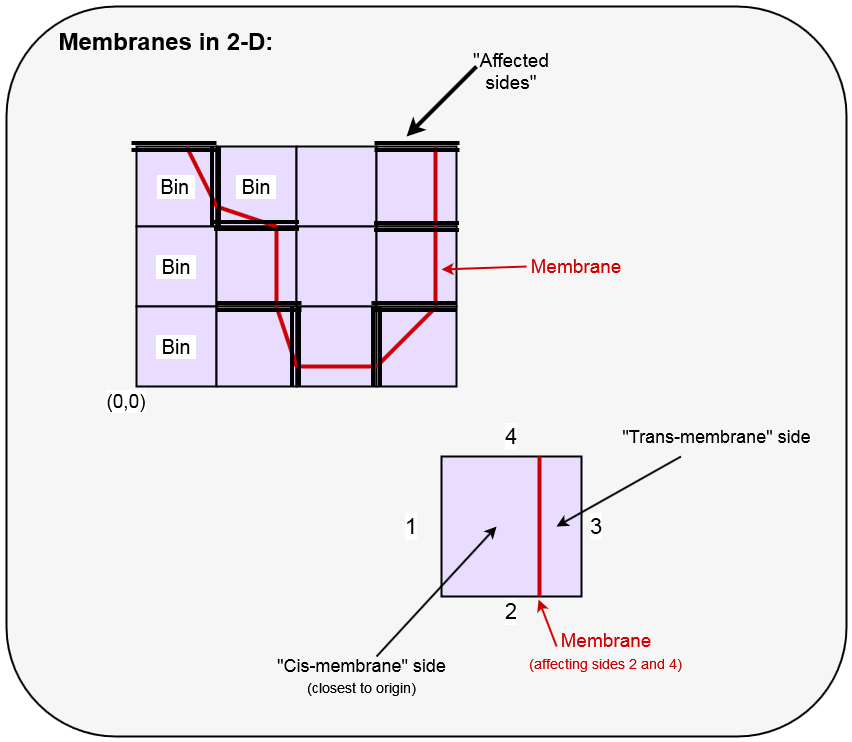
- Membranes divide cells in exactly 2 (generally unequal) parts, referred to as "Cis-side" and "Trans-side" of the membrane. This will hold in both 1D, 2D and 3D.
The terms "cis" and "trans" refer to the relative position with respect to the coordinate origin of the system.
(For historical reasons, in the software, "Cis" is sometimes referred to as "the regular side", while "trans" as "the other side" or "B side".) - The relative sizes of the "cis" and "trans" sides are recorded, and are used to estimate distances for "diffusion" processes across membranes.
- In 1-D, the membranes are assumed not to touch any edge; in 2-D, they touch exactly 2 edges; in 3-D, they touch 3 or 4 faces.
- At present, the membranes are in fixed positions and don't vary with time; future iterations will probably simulate drift, invagination, fusion, etc.
- "Compartments" are not actively modeled, at least for now. Compartments are taken to be an emergent property of regions sorrounded by membranes (or "sandwiched" by them, in the case of 1-D)

Details of the data structure
In 1-D, in the absence of membranes, the concentrations of each of the chemical species in each of the bins is a 2-dimensional NumPy array (matrix) of floating-point numbers,
of dimensions (n_species x n_bins). That's stored in the BioSim1D class variable system.
At least for now, starting with the Beta 13 release, that same matrix is used for the concentrations values in membrane-free bins, as well as for the "cis" sides
of bins with membranes.
The "trans" sides use their own 2-dimensional NumPy array, currently named system_B; that will generally be a sparse matrix.
This early implementation is tentative and subject to change. However, the API will largely or entirely protect higher-level code, as well as the end user, from
the underlying data structure details.