User Guide
A basic knowledge of Python (and, optionally, of Jupyter Notebooks) is all that is required to run "in-silico experiments" (simulations) with Life123 !
NOTE: for examples of usage and tutorials, see the Experiments.
The unit tests of the various python functions can also be of help.
For the JavaScript modules, see the Visualization section.
How to get started using these libraries: see our quick start page!
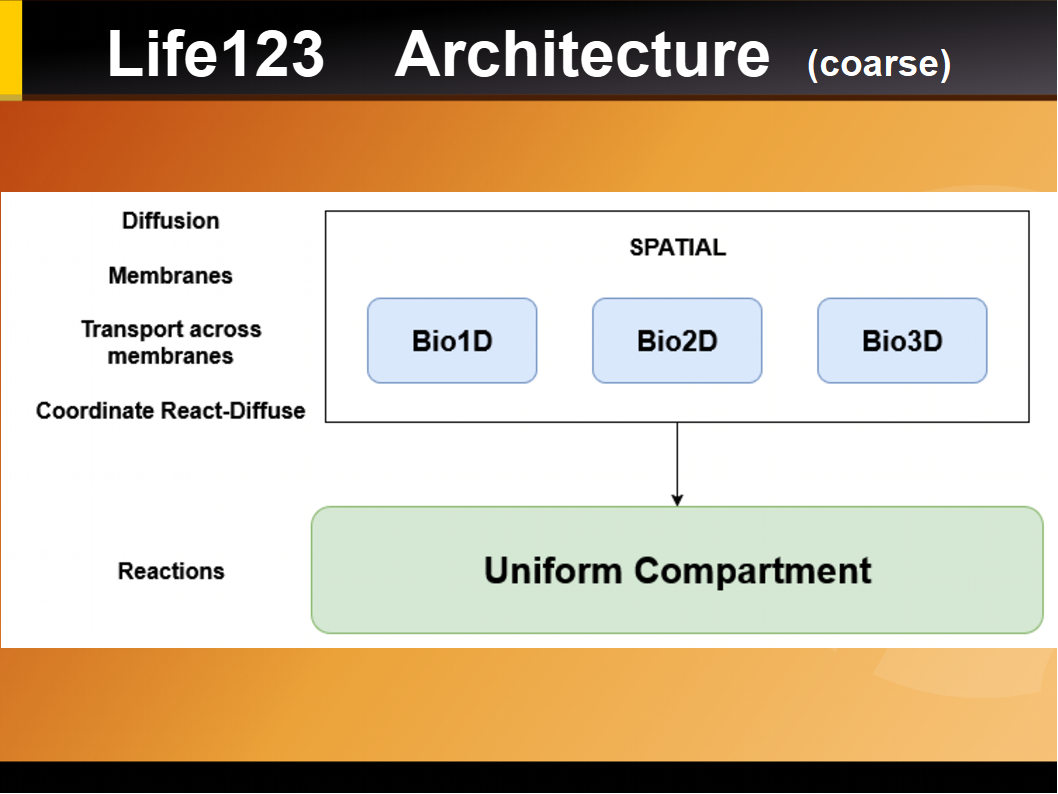
The following libraries (python classes) are being used:
CORE CLASSES (Simulations)
BioSim1D
1D simulations of diffusion and reactions, including passive transport across membranes.
BioSim2D
2D simulations of diffusion and reactions. Early implementation of membranes.
BioSim3
3D simulations of diffusion and reactions (early implementation)
ChemData
Data about all the chemicals and (if applicable) reactions,
including:
- names
- diffusion rates
- macro-molecules Binding Site Affinities (for Transcription Factors)
Notes: * for now, the temperature is assumed constant everywhere, and unvarying (or very slowly varying)
* this class contains (extends) the following other classes: ChemCore, Diffusion, Macromolecules
Numerical
Assorted, general numerical methods
Reactions - Various Subtypes
7 classes:
ReactionCommon, ReactionOneStep,
ReactionUnimolecular, ReactionSynthesis, ReactionDecomposition,
ReactionEnzyme, ReactionGeneric
The objects from those classes are managed by the class ReactionRegistry.
ReactionRegistry
Manage the reaction-specific classes, such as ReactionUnimolecular, ReactionSynthesis, ReactionDecomposition, ReactionGeneric, ReactionEnzyme, etc. (this class was formerly called "Reactions")
ReactionKinetics / VariableTimeSteps
2 classes:
1) Static methods about reactions kinetics
2) Methods for managing variable time steps during reactions
ThermoDynamics
Manage the Thermodynamics aspects of reactions:
changes in Gibbs Free Energy, Enthalpy, Entropy - and how
they relate to equilibrium constant, at a given temperature.
This class does NOT get instantiated
"K" (equilibrium constant - from either kinetic or thermodynamic data;
if both present, they must match up!)
"delta_H" (change in Enthalpy: Enthalpy of Products - Enthalpy of Reactants)
"delta_S" (change in Entropy)
"delta_G" (change in Gibbs Free Energy)
Note - at constant temperature T :
Delta_G = Delta_H - T * Delta_S
Equilibrium constant = exp(-Delta_G / RT)
UniformCompartment
Used to simulate the dynamics of reactions (in a single compartment)
VISUALIZATION LIBRARIES
Colors
For color-related matters
PlotlyHelper
To assist in the use of the plotly library
PyGraphVisual
Facilitate data preparation for graph visualization using the Cytoscape.js library. The development of this library is shared with the sister project BrainAnnex.org
LOGGING LIBRARIES
GraphicLog
To simplify use of HtmlLog and Vue components from within this project
HtmlLog
An HTML logger to file, plus optional plain-text printing to standard output
UTILITY LIBRARIES
CollectionTabular
A "tabular collection" is a Pandas dataframe
built up from a sequence of "snapshots" of data that's in the form of a python dictionary
(representing a list of values and their corresponding names),
such as the state of the system or of parts thereof.
Each data "snapshots" is taken at different times,
or results from varying some parameter.
Each snapshot - incl. its parameter values and optional captions -
will constitute a "row" in a tabular format
MAIN DATA STRUCTURE for "tabular" collections:
A Pandas dataframe
CollectionArray
Use this structure if your "snapshots" (data to add to the cumulative collection) are Numpy arrays,
of any dimension - but always retaining that same dimension.
Usually, the snapshots will be dump of the entire system state, or parts thereof, but could be anything.
Typically, each snapshot is taken at a different time (for example, to create a history), but could also
be the result of varying some parameter(s)
DATA STRUCTURE:
A Numpy array of 1 dimension larger than that of the snapshots
EXAMPLE: if the snapshots are the 1-d numpy arrays [1., 2., 3.] and [10., 20., 30.]
then the internal structure will be the matrix
[[1., 2., 3.],
[10., 20., 30.]]
Collection
A "Collection" is a list of snapshots of any values that the user wants to preserve,
such as the state of the entire system, or of parts thereof,
either taken at different times,
or resulting from varying some parameter(s)
This class accept data in arbitrary formats
MAIN DATA STRUCTURE:
A list of triplets.
Each triplet is of the form (parameter value, caption, snapshot_data)
1) The "parameter" is typically time, but could be anything.
(a descriptive meaning of this parameter is stored in the object attribute "parameter_name")
2) "snapshot_data" can be anything of interest, typically a clone of some data element.
3) "caption" is just a string with an optional label.
If the "parameter" is time, it's assumed to be in increasing order
EXAMPLE:
[
(0., DATA_STRUCTURE_1, "Initial state"),
(8., DATA_STRUCTURE_2, "State immediately after injection of 2nd reagent")
]
Diagnostics
For the management of reaction diagnostic data
HistoryBinConcentration
For the management of historical data for bin concentrations
HistoryReactionRate
For the management of historical reaction-rate data for Uniform Compartments
HistoryUniformConcentration
For the management of historical concentration data for Uniform Compartments