|
Date
(MOST RECENT at top)
|
New Additions & Bug Fixes
(all main branch unless otherwise specified) |
|
Sep. 4, 2025
1.0 Release Candidate 6
(RC5 not released)
|
PRESENTATION / DOCUMENTATION
- Talk about Life123 given at the
"Aging & Gerontology 2025" conference in San Francisco,
on the topic "Dynamical Modeling as a Precision Tools for Longevity Science"

- Blog entry based on the talks given at the above conference and the previous one
- Improvements in the UX of the filter selector in the Experiments page
- A locally-hosted version of the "NB Viewer" web app (to render archived Jupyter notebooks), as a replacement for the
website
nbviewer.org, which is periodically afflicted by overloads
- Expansion and re-organization of the home page
HIGHLIGHTS / CORE / NEW FEATURES
- Native support for enzymatic reactions
- Major restructuring of the class
BioSim1D
- Early implementation of membranes in 2D
EXPERIMENTS (listing of all experiments)
- New 1D diffusion experiments:
diffusion\localized_transient, diffusion\membrane_gradient_diffusion_1
- New 1D reaction-diffusion experiments:
reaction_diffusion\transient_getting_mopped_up,
reaction_diffusion\membranes_racing_condition_1
- New experiment
reactions_single_compartment\enzyme_1_a_technical
- Overhauled experiment
reactions_single_compartment\enzyme_1_a
- Updated experiments
reactions_single_compartment\react_2_a, react_2_d
- Tweaks to experiment
1D\diffusion\diffusion_3
- Updated numerous 1D experiments
MAIN INFRASTRUCTURE
- Dropped dependencies on the
neoaccess and neo4j libraries
SUPPORTING LIBRARIES
- BioSim1D (and derived classes) :
Split into multiple parts: a base (parent) class System1D, which BioSim1D now extends.
Some of the methods got moved into new separate classes: Membranes1D and Diffusion1D
New methods: get_system_time(), total_chem_mass(), snapshot_system()
Simplification to reaction_step()
visualize_system() now also shows membranes, if present
Added optional argument `reaction` to class constructor
Added argument `max_points` to plot_history_single_bin()
Added argument `plot_bg_color` to visualize_system()
Added arguments `max_amplitude` and `clip` to inject_bell_curve(), and introduced alternate argument name `center` for `mean`
Added arguments `show_status` and `fraction_max_step` to diffuse() ; `time_step` now returned in status
Added arguments `fraction_max_step` and `show_status` to react_diffuse()
plot_history_single_bin() now also accepts lists as argument `title_prefix`
Renamed argument `species_label` to `chem_label` in bin_concentration()
Dropped argument `snapshots` from react()
Bugfixes to describe_state(), when membranes are involved
- UniformCompartment :
New methods reaction_step_common_fixed_step(), _fetch_concs_for_rnx()
Taken out the inner loop of _reaction_elemental_step() into separate method _inner_reaction_loop()
Dropped methods compute_all_reaction_rates(), _reaction_elemental_step_SINGLE_REACTION()
Dropped argument `verbose` from is_in_equilibrium()
Dropped argument `rxn_index` from _inner_reaction_loop()
Better exception handling in reaction_step_common_fixed_step()
Changes to single_compartment_react() : reactions with fixed time steps no longer attempt to reduce step size
Enhanced ExcessiveTimeStepHard Exception details
In _inner_reaction_loop() and other functions, replaced `increment_vector_single_rxn` with `increment_dict_single_rxn`
The validation check for negative concentrations is now done by the calling function _reaction_elemental_step()
Relocated `temperature` as an object variable of the class; added `temp` argument to class constructor
- Diffusion :
New method get_max_diffusion_rate()
Renamed argument `label` to `chem_label` in set_diffusion_rate()
- Various Reaction classes :
New classes ReactionCommon, ReactionElementary,
ReactionUnimolecular, ReactionSynthesis, ReactionDecomposition
Changes to classes ReactionGeneric and ReactionEnz (now renamed ReactionEnzyme),
to make use of the new classes above
Introduced ReactionEnzyme as a reaction_type
New method add_reaction_from_object()
Renamed method extract_reactant_names() to extract_reactant_labels(), and other similar changes
Added argument `sort_by_index` to labels_of_active_chemicals()
Dropped argument `exclude_enzyme` from methods extract_reactant_names(), extract_product_names() and extract_chemicals_in_reaction()
- ReactionKinetics :
New method compute_reaction_rate_elementary()
New method compute_reaction_quotient() in this class, and a series of reaction_quotient() methods
for each of the various specific reaction classes
Renamed compute_reaction_rate_elementary() to compute_reaction_rate_first_order()
Added support for irreversible reactions to compute_reaction_rate()
Changes to _inner_reaction_loop() and _reaction_elemental_step(),
to use `conc_dict ` instead of `conc_array` and `name_mapping`
- History :
Added argument `downsize` to HistoryBinConcentration.bin_history() ;
massive efficiency improvement to that function
Bugfix to _diagnose_history_problem()
- Diagnostics :
Adjustments to save_rxn_data() from recent changes
Support for multiple reaction rates: in Diagnostics.save_rxn_data() , ReactionEnzyme.step_simulation()
and UniformCompartment.capture_rate_snapshot()
- Early implementation of new classes
System2D and Membranes2D
VISUALIZATION
- Added argument `modify` to
PlotlyHelper.combine_plots()
- New method
assemble_title()
|
|
May 31, 2025
1.0 Release Candidate 4
(RC3 not released)
|
HIGHLIGHTS / CORE / NEW FEATURES
- Overhaul of implementation of membranes in 1D, including their visualization
- Simulation of passive transport across membranes in 1D
- Visualization: stacks of heatmaps in 1D (one per chemical)
- Substantial improvement of name consistency for functions and arguments
EXPERIMENTS (listing of all experiments)
- New 1D experiments:
diffusion\diffusion_3,
diffusion\membranes_1, diffusion\membranes_2, diffusion\membranes_3, reaction_diffusion\rd_2
- New experiment:
visualization\plotly_helper\heatmap_stack_1D
- Updated 1D experiments:
diffusion\overly_large_single_timesteps,
diffusion\diffusion_1 , diffusion\diffusion_2, 1D\reaction_diffusion\rd_1
- Update to experiment
reactions_single_compartment\react_1_a
MAIN INFRASTRUCTURE
- All recently-added classes are now included in
__init__.py
- Upgraded the version of several external libraries: jupyterlab, jupytext, plotly, numpy, scipy, pandas, jsonschema
SUPPORTING LIBRARIES
- BioSim1D :
Introduction of passive membrane transport to _diffuse_step_single_chem_3_1_stencil()
New methods: membranes_list(), membrane_on_left(), membrane_on_right(), set_permeability(), change_permeability(), chem_quantity()
Obsoleted and eliminated the old membrane implementation (several methods affected)
Overhauled methods set_membranes() and system_heatmaps()
The increment_vector array is now passed as argument to the various "_diffuse_step_single_chem_" methods
Cleanup and simplifications in diffuse_step()
Change in return value of diffuse()
New arguments: `vertical_lines_to_add` added to plot_history_single_bin(), `caption` to enable_history(),
`include_captions` to get_bin_history(), `show` to visualize_system(), `title_prefix` to plot_history_single_bin()
Renamed argument `species_name` to `chem_name` in inject_gradient()
NAME CONSISTENCY: Renamed arguments `species_name` to `chem_label` , and `species_index` to `chem_index`
in numerous class methods
- ChemData and related classes:
Change in ChemData's instantiation: default chemical names are auto-generated when chemical names not provided
Renamed ChemData.assert_valid_species_index() to assert_valid_chem_index()
Renamed argument `species_index` to `chem_index` in ChemCore.get_label()
Renamed argument `species_index` to `chem_index` in Diffusion.get_diffusion_rate()
- Numerical :
Modifications to gradient_order4(), to conform to new Numpy version
- History :
New method is_enabled()
VISUALIZATION
- New method
PlotlyHelper.heatmap_stack_1D()
- Added argument `log_y` to
PlotlyHelper.plot_pandas() and argument `layout_index` to combine_plots()
DOCUMENTATION
DEPLOYMENT
- Added "pyproject.toml" to the repo
BUG FIX
- Bugfix to
BioSim1D.system_heatmap()
|
|
Jan. 25, 2025
1.0 Release Candidate 2
|
HIGHLIGHTS / CORE / NEW FEATURES
- Support for reaction-diffusion in 2D
- Extensive expansion of 2D experiments
- Visualization: new heatmaps for 2D systems, and improved/simplified heatmaps in 1D
- Streamlined support for capture and visualization of bin concentration history in 1D and 2D
- Support for associating colors to individual chemicals, and automatically using those registered colors in visualizations
EXPERIMENTS (listing of all experiments)
- New 2D experiments:
2D/diffusion/diffusion_2a, 2D/diffusion/diffusion_2b, 2D/reaction_diffusion/rd_1
- New 1D experiment:
1D/diffusion/diffusion_2
- Extensive simplification and extension of experiments
2D/diffusion/diffusion_1, 1D/reaction_diffusion/rd_1
- Additional work on experiment
reactions_single_compartment/enzyme_3
- Updated experiments in "reactions_single_compartment":
enzyme_1_a, enzyme_1_b, enzyme_1_c, up_regulate_1
- Updated 1D experiments:
1D/reaction/reaction_1, 1D/reaction/reaction_8, 1D/diffusion/diffusion_1
- Updated all 2D experiments with the new instantiation arguments of the class
BioSim2D
MAIN INFRASTRUCTURE
- Deprecated the old implementation of 1-D membranes
- Introduced a new History module to simplify/streamline history keeping in bin simulations in 1D and 2D, as well as in UniformCompartment simulations
SUPPORTING LIBRARIES
- BioSim1D :
Changes to class constructor: argument "nbins" is now required
Introduced use of new History module
Added history keeping for reaction-diffusion in 1D
New methods: get_reaction_handler(), get_chem_data(), reaction_in_equilibrium(), get_reactions(), selected_concentrations(), enable_history(), capture_snapshot(), get_bin_history(), plot_history_single_bin(), system_snapshot_arr(), check_mass_conservation(), heatmap_single_chem()
Added argument "chem_label" to inject_conc_to_bin(), and renamed argument "species_index" to "chem_index"
Added argument "smoothed" to plot_history_single_bin()
Renamed argument "caption" to "title_prefix" in visualize_system()
Renamed argument "species_index" to "chem_index" in inject_conc_to_bin()
Enhancements to method describe_state()
Changes to color handling in visualize_system() and system_heatmap(). Added support for pre-assigned colors in line_plot()
Registered chemical colors are now respected in visualize_system() and in plot_history_single_bin()
Expanded heatmap_single_chem(), now renamed system_heatmap(); ditched the old version of system_heatmap()
More validation in various class methods
Deprecated the old implementation of 1-D membranes
- BioSim2D :
Introduced support for reaction-diffusion
Changes to class constructor: argument "nbins" is now required
Introduced use of new History module
Added history keeping for reactions and reaction-diffusion in 2D
New methods: get_reactions(), assert_valid_bin(), selected_concentrations(), get_system_time(), capture_snapshot(), react_diffuse(),
system_snapshot_array(), check_mass_conservation(), heatmap_single_chem(), heatmap(), system_heatmaps(), plot_history_single_bin()
Method heatmap() renamed heatmap_single_chem_greyscale()
Dropped unused argument "snapshots" from react()
Change in arguments to set_bin_conc() and set_bin_conc_all_species()
Renamed argument of enable_history() from "labels" to "chem_labels"
Added argument "smoothed" to plot_history_single_bin()
In system_snapshot(), argument "species_index" renamed to "chem_index", and added argument "chem_label"
In set_bin_conc(), added argument "chem_label", and dropped "species_index"
Changes to system_snapshot_array(), renamed system_snapshot_arr_xy(); in particular, transposing the matrix to obtain a Cartesian grid
Changes to selected_concentrations()
More validation, and a default "title" argument value, to plot_history_single_bin()
Bugfix to heatmap_single_chem()
Support for registered colors in plot_history_single_bin() and in system_heatmaps()
Added bin-number validations across the class
- PlotlyHelper :
Relocated the color-related method get_default_colors() to the new class "Colors"
Added argument "smoothed" to plot_pandas()
New methods: heatmap_grid(), _optimal_subplot_grid_size()
- Colors :
New class "Colors"; all color-related methods, including some formerly in the class "PlotlyHelper" are being located here
Renamed get_default_colors() to assign_default_colors()
New methods: get_named_colors_mapping(), to_rgb(), lighten_color(), assign_default_heatmap_colors()
- ChemData (ChemCore) :
New methods: get_registered_colors(), get_color_mapping_by_label(), get_color_mapping_by_index(), get_all_colors()
- History :
New class "History" and its 3 derived (children) classes: "HistoryBinConcentration", "HistoryReactionRate" and "HistoryUniformConcentration"
Step-count values are now saved in the histories
- UniformCompartment :
This class now uses the new "History" module to save the rate history
New methods: enable_history(), capture_rate_snapshot(), capture_conc_snapshot()
Dropped argument "snapshots" of single_compartment_react()
Dropped the method add_snapshot()
Completely re-implemented the method add_rate_to_conc_history()
- Diagnostics :
Ditched method get_system_history_with_rxn_rates()
Several bugfixes in class Diagnostics
VISUALIZATION
- Introduced 2D heatmaps, with a "Cartesian" view (y-axis pointing up)
- Added argument "cartesian" to various methods of class "BioSim2D":
system_snapshot(), describe_state(), heatmap_single_chem_greyscale(), heatmap_single_chem(), system_heatmaps()
- Added argument "cartesian" to
PlotlyHelper.heatmap_grid()
- Improved/simplified heatmaps in 1D
- Streamlined visualization of bin concentration history in 1D and 2D
DOCUMENTATION
DEPLOYMENT
- Added file "experiments\2D\reaction_diffusion\set_path.py"
BUG FIX
- Corrections to
BioSim2D.heatmap_single_chem()
- Several bugfixes in class Diagnostics
- Corrections to "environment.yml" (used for deployment to mybinder.org)
|
|
Dec. 16, 2024
1.0 Release Candidate 1
|
HIGHLIGHTS / CORE / NEW FEATURES
- Added support for attaching plotting colors to chemicals (primarily affecting classes "ChemData" and "Reactions")
EXPERIMENTS (listing of all experiments)
- Split and expanded experiment "enzyme_2" into "enzyme_2_a" and "enzyme_2_b"
- Enhancenents and simplification to "enzyme_4"
- Re-classified the 1D version of "state_space_1"
- Updated several older experiments to the new library version
MAIN INFRASTRUCTURE
- Separated the "AllReactions" class out of the "ChemData" class, renamed it "Reactions" and moved it to the file reaction.py
- Renamed the file reaction.py to reactions.py
- Renamed the class "Reaction" (singular) to "ReactionGeneric"
SUPPORTING LIBRARIES
- UniformCompartment :
Added new argument, "reactions", to the class constructor
Added 2 methods get_reactions() and get_single_reaction()
- ChemData :
Overhauled the class constructor, and added arguments "labels" and "plot_colors"
Dropped method init_chemical_data()
Added more accepted data types to assert_valid_diffusion()
- Reactions :
Renamed names_of_active_chemicals() to labels_of_active_chemicals()
- BioSim1D :
In method bin_concentration() renamed argument "species_name" to "species_label"
Simplifications to class constructor; renamed argument "reactions" to "reaction_handler"
- BioSim2D :
Simplifications to class constructor; renamed argument "reactions" to "reaction_handler"
VISUALIZATION
- Enhancements to
PlotlyHelper.plot_pandas(), to deal with missing individual curve colors
- New method
ChemCore.get_plot_color()
ChemCore.all_chemicals() now also includes column of assigned plot colors
- Changes to
UniformCompartment.plot_history()
|
|
Nov. 18, 2024
1.0 Release Candidate 0
|
HIGHLIGHTS / CORE / NEW FEATURES
- Optional automatic export of reaction data (system state) to a CSV log file
- Enhanced support for enzymatic reactions
EXPERIMENTS (listing of all experiments)
- Expanded experiment
reactions_single_compartment/react_2, and renamed it react_1_a
- New experiments
react_1_b, react_1_c, enzyme_2 and enzyme_3
- Revised experiments
cascade_2_a, cascade_2_b, cascade_2_c, react_2_a, react_2_b, react_2_c, react_2_d, mystery_reaction_1
- Updated several experiment to the latest libraries version
MAIN INFRASTRUCTURE
- Reaction rates are now "first-class citizens", stored alongside historical concentration data,
rather then being relegated to being part of diagnostic data
SUPPORTING LIBRARIES
- UniformCompartment :
Added argument "enable_diagnostics" to class constructor
Various enhancements to single_compartment_react(): better status reporting; added argument "report_interval",
and dropped obsolete argument "reaction_duration"; reaction rates are now automatically stored
New methods start_csv_log(), add_rate_to_conc_history() and get_rate_history()
Enhancements to set_conc()
Changes to add_snapshot()
- ReactionKinetics :
Expanded method estimate_rate_constants_simple()
- Reaction :
Changed name of rate-constant arguments in class constructor
- Diagnostics :
New method get_system_history_with_rxn_rates()
Added argument "caption" to save_diagnostic_conc_data()
- Renamed class "MovieTabular" to "CollectionTabular", "MovieArray" to "CollectionArray", and "MovieGeneral" to "Collection"
- CollectionTabular :
New method update_last_snapshot()
New argument "return_copy" to get_dataframe()
Enhancements to method store()
- New class "ReactionEnz"
VISUALIZATION
- New method
PlotlyHelper.combine_in_vertical_grid()
- Tweaks to
plot_pandas(), including additional values allowed in argument "vertical_lines_to_add"
- Tweaks to
UniformCompartment.plot_history()
DOCUMENTATION
BUG FIX
- Bugfix to
UniformCompartment.single_compartment_react()
|
Beta versions:
|
|
Oct. 11, 2024
1.0 Beta 39
|
CORE / NEW FEATURES
- Added support for irreversible reactions : changes to classes
UniformCompartment and Reaction
EXPERIMENTS (listing of all experiments)
- New experiments
reactions_single_compartment/enzyme_1 and react_2_d
- Renamed experiments
enzyme_1, enzyme_2, enzyme_3 to, respectively, enzyme_hypothetical_1, enzyme_hypothetical_2, enzyme_hypothetical_3
- Updated several experiments to this new library version
MAIN INFRASTRUCTURE
- "labels", rather than "names" now index chemicals; by default, "labels" are equal to the given "names"
- Renamed class
ReactionDynamics to ReactionKinetics, and renamed file "reaction_dynamics.py" to "reaction_kinetics.py"
SUPPORTING LIBRARIES
- UniformCompartment :
Changed the defaults for the argument "snapshots" in single_compartment_react()
Obsoleted set_diagnostics()
- ReactionKinetics :
New method exact_solution_unimolecular_irreversible()
Moved method UniformCompartment.estimate_rate_constants_simple() and estimate_rate_constants_association() to the class ReactionKinetics; also moved compute_reaction_rate(), and added 2 arguments to it
Renamed exact_solution_combination_rxn() to exact_solution_synthesis_rxn() , and ._exact_solution() to exact_solution_unimolecular_reversible()
- ChemCore :
New method get_name_mapping()
Numerous changes to use "labels", rather than "names", to index chemicals
- Diagnostics :
Added "aborted" column to the individual-reaction diagnostic dataframes
New method get_rxn_rates()
Renamed get_diagnostic_decisions_data() to get_decisions_data(),
and get_diagnostic_rxn_data() to get_rxn_data()
VISUALIZATION
- New argument, "range_y" added to
UniformCompartment.plot_history()
and to PlotlyHelper.plot_pandas(); renamed some args in plot_history()
- Renamed some args in
PlotlyHelper.plot_curves()
- Renamed some args in
plot_pandas(); and added "x_label" argument
- New method
MovieTabular.set_field_last_snapshot()
BUG FIX
- Bugfix in
PlotlyHelper.plot_curves()
DOCUMENTATION
|
|
Sept. 8, 2024
Beta 38
|
EXPERIMENTS (listing of all experiments)
- Updated experiments
reactions_single_compartment/react_2_b
- Completed new experiment
react_3_b
- Expanded experiment
react_2_c
- New experiment
mystery_reaction_2.py
- Simplified experiment
mystery_reaction_1
MAIN INFRASTRUCTURE
- Renamed class "ReactionDynamics" to "VariableTimeSteps"
- New class "ReactionDynamics"
SUPPORTING LIBRARIES
- UniformCompartment :
New methods get_chem_data() and estimate_rate_constants_association()
The argument "columns" of get_history() is also allowed to be a string
Renamed compute_reaction_delta_rate() to compute_reaction_rate()
- ReactionDynamics :
New methods approx_solution_combination_rxn() and exact_solution_combination_rxn()
Moved method solve_exactly() from class "UniformCompartment" to "ReactionDynamics"
Absorbed the class "Heuristics" into "ReactionDynamics"
Tweaks to ._exact_solution()
- Numerical :
New methods two_vector_least_square() and simple_least_square()
VISUALIZATION
- Relocated method
UniformCompartment.plot_data() as PlotlyHelper.plot_pandas()
- Added argument "legend_header"
to plot_data()
|
|
July 25, 2024
Beta 37
|
EXPERIMENTS (listing of all experiments)
- Expanded experiments
react_2_a and react_2_b
- Updated experiments
react_1, react_2_c and enzyme_4
- Simplified the export of logs with the library
ipynbname
- Started introducing notebook tips about dealing with failed python imports
MAIN INFRASTRUCTURE
- Reaction rates are now saved alongside other diagnostic data
- Diagnostics-related parts of class
UniformCompartment moved to a separate class, Diagnostics, in "diagnostics.py"
- Chemicals passed to
add_reaction(), in various modules, no longer have to be pre-registered
- Added new function
check_version() to file "__init__.py"
SUPPORTING LIBRARIES
- ReactionDynamics :
Renamed class RxnDynamics to ReactionDynamics, and moved it into a separate file, "reaction_dynamics.py";
moved into it several object variables and methods from the class UniformCompartment
Ditched un-necessary fields formerly stored in the diagnostic history of individual reactions
- UniformCompartment :
Tweaks to plot_history()
Changes to compute_all_reaction_rates(), formerly named compute_all_reaction_deltas()
Renamed set_diagnostics() to enable_diagnostics()
- AllReactions :
New method get_chemicals_indexes_in_reaction()
VISUALIZATION
- New method
UniformCompartment.plot_data()
DOCUMENTATION
- Expansion and restructuring of the Reactions page
- Started added categorization tags to the notebooks listed in the Experiments page
- Fixed broken repository links in the Guide (from the major project restructuring of the previous release)
DEPLOYMENT
- Set version
jsonschema==4.17.3 in requirements.txt
- Upgraded version of the
neoaccess library to 4.4.1
- Dropped requirements for libraries
ipywidgets and jupyter-dash
- Tweaks to "quick.bat", the batch file to start JupyterLab
|
|
June 23, 2024
Beta 36
(Beta 34 and 35 not released)
|
CORE / NEW FEATURES
- Life123 is now available from the standard PyPI release :
pip install life123
EXPERIMENTS (listing of all experiments)
- Updated all experiments, to reflect the new import names
MAIN INFRASTRUCTURE
- Major overhaul of file structure, readied for PyPI release
SUPPORTING LIBRARIES
- Ditched obsolete class "ReactionDynamics" and obsolete method
UniformCompartment.curve_intersection()
VISUALIZATION
- Ditched
matplotlib and a test notebook using it. (Note: plotly has long been used instead.)
DEPLOYMENT
- Faulty releases Beta34 and 35 were withdrawn (PyPI release is the same for 35 and 36)
DOCUMENTATION
|
|
June 15, 2024
Beta 33
|
CORE / NEW FEATURES
- Improved management of variable time steps for networks of reactions,
employing new norms (measures of change/imminent onset of instability), more presets, and better integration of the norm values
EXPERIMENTS (listing of all experiments)
- Updated a large number of experiments, to reflect all code changes
- Variable-timestep presets are now used in all experiments that previously set individual low-level parameters
MAIN INFRASTRUCTURE
- Renamed class
ReactionDynamics to UniformCompartment
SUPPORTING LIBRARIES
- UniformCompartment (formerly named ReactionDynamics) :
Taken out several methods, and moved them to new separate static class, "RxnDynamics"
New methods update_thresholds() and delete_thresholds()
For variable time steps, added several new presets and two new norms (measures of change), norm_C and norm_D
Previous system state at tn-1 is now kept, and used by the new norms
Norm usage is now stored and displayed
Changes in the return value of adjust_timestep(), and other enhancements
Absorbed the "error_abort_step_factor" class property into the "step_factors" dict class property
In single_compartment_react(), argument name change from "reaction_duration" to "duration", and
changed argument "explain_variable_steps" from Boolean to pair (start-end time)
Tweaks to output when using argument "explain_variable_steps"
Replaced the method curve_intersection() with the new curve_intersect()
Tweaks to plot_history()
Ditched obsolete criterion_fast_reaction()
- AllReactions:
New method number_of_active_chemicals()
DOCUMENTATION
BUG FIX
- Bugfix, and additional validation, in
set_thresholds()
|
|
May 31, 2024
Beta 32
|
CORE / NEW FEATURES
- Implementation of additional termination criteria for reaction simulations
EXPERIMENTS (listing of all experiments)
MAIN INFRASTRUCTURE
- Added file
src/__init__.py
SUPPORTING LIBRARIES
- ReactionDynamics
Added argument "preset" in class constructor; deprecated argument "shared"
Added arguments "max_steps" and "stop" to single_compartment_react();
renamed argument "reaction_duration" to "duration"
Added new "slower" preset to use_adaptive_preset()
Changes to set_conc()
Small correction to display_thresholds()
Streamlined method curve_intersection(), now renamed curve_intersect()
New argument "verbose" to is_in_equilibrium() ; tweaks to reaction_in_equilibrium()
- ChemData:
Light re-organization
Enforcement of unique chemical names
New method name_exists()
Added argument "name" to Diffusion.get_diffusion_rate()
Renamed argument "diffusion_rate" to "diff_rate" in add_chemical_with_diffusion()
New methods AllReactions.inactivate_reaction(), active_reaction_indices(), indexes_of_active_chemicals()
Added argument "include_inactive" to number_of_reactions()
- Reaction:
Major change in internal data structure: chemical names now used instead of their indices;
the "AllReactions" class is no longer passed during object instantiation
New methods extract_reactant_names() and extract_product_names()
Enhancement to reaction_quotient()
- Numerical:
Extended curve_intersect_interpolate()
New method reach_threshold()
Overhauled the implementation of curve_intersect_interpolate(), now renamed curve_intersect()
DOCUMENTATION
- Updated the affected Reference Guides
- Misc. enhancements and problem fixes on the website
BUG FIX
- Bugfix to
AllReactions.clear_reactions_data()
- Bugfix to
ReactionDynamics.plot_history()
|
|
May 7, 2024
Beta 31
|
CORE / NEW FEATURES
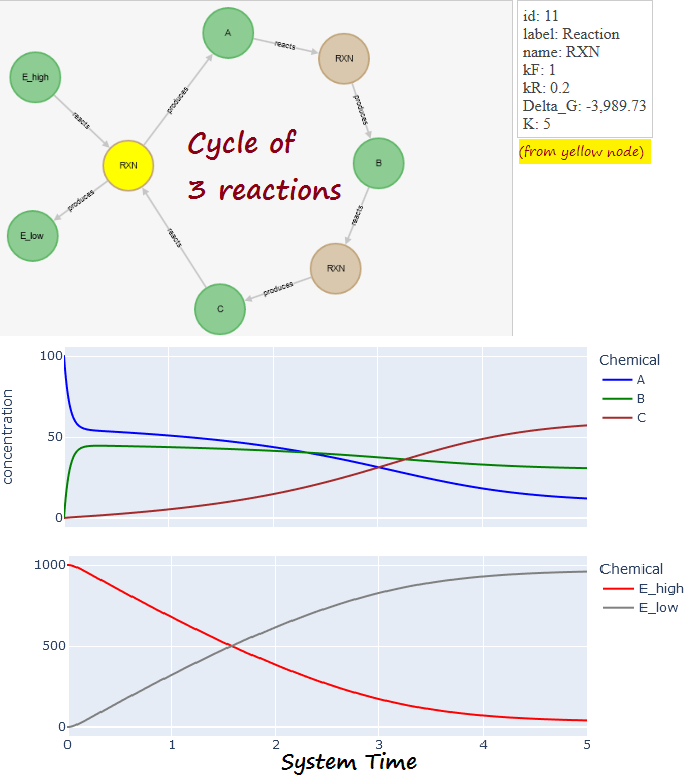
- Improved visualization of networks of reactions (details below). Development has been shared with sister project BrainAnnex.org
EXPERIMENTS
- Updated all experiments affected by the changes
MAIN INFRASTRUCTURE
SUPPORTING LIBRARIES
- Added
utilities\comparisons.py
VISUALIZATION
- Enhanced visualization of networks of reactions, using Vue component
vue_cytoscape_2
- New class
PyGraphVisual
- New method
HtmlLog.export_plot_Vue_unpack_args()
- New argument "unpack" to
GraphicLog.export_plot()
- New method
ReactionDynamics.plot_reaction_network()
- Changes to
ChemData.prepare_graph_network()
DOCUMENTATION
BUG FIX
- In network visualizations, corrected issue of some reactions parameters being incorrectly shown on nodes of graph structure, when they should have appeared on edges
- Corrected relative paths in
src\modules\visualization\graphic_log.py
EXAMPLE. From experiment
A cycle of reactions (CLICK ON IMAGE BELOW FOR INTERACTIVE VERSION):
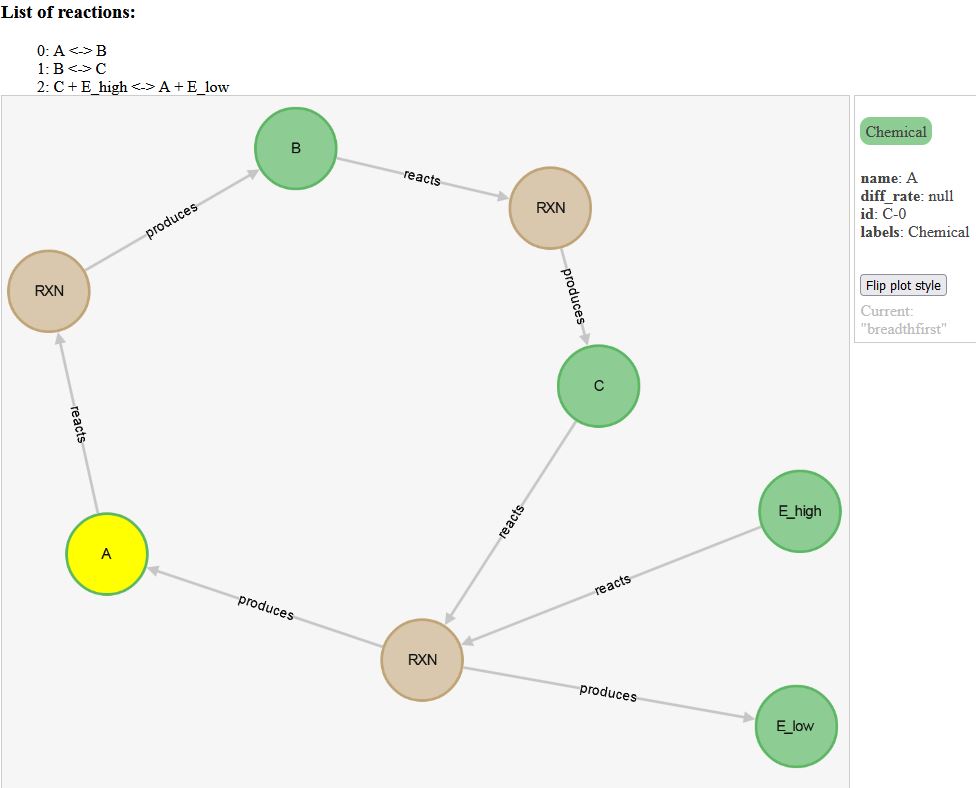
|
|
Dec. 9, 2023
Beta 30.1
|
CORE / NEW FEATURES
- Streamlined and simplified the programming interface
- Added more data-analysis tools
EXPERIMENTS
MAIN INFRASTRUCTURE
- For single-compartment reactions, the new default is variable adaptive time steps
- Changed default reaction orders to the stoichiometry coefficients
SUPPORTING LIBRARIES
- ReactionDynamics:
New methods estimate_rate_constants(), find_equilibrium_conc(), solve_exactly(),
number_of_reactions(), add_reaction(), describe_reactions(), prepare_graph_network(),
show_adaptive_parameters(), use_adaptive_preset()
Added "columns" argument to get_history()
Simplified reaction_in_equilibrium(); added handling of pathological cases
Improvements to stoichiometry_checker() ; dropped argument "suppress_warning"
Enhancements to set_single_conc()
Added arguments "names" and "shared" to the class constructor
Cleanup of obsolete code; adjustments to some default values
- Reaction:
New methods extract_equilibrium_constant(), reaction_quotient()
- ChemData:
Change in value returned by add_reaction()
- Numerical:
New methods gradient_uneven_grid(), _finite_diff_weights()
Tweaks to _compute_gradient() , now renamed _compute_derivative()
VISUALIZATION
- New class "PlotlyHelper", with methods
get_default_colors(), plot_curves(), combine_plots();
- Using this new class, streamlined
ReactionDynamics.plot_curves(), and renamed it plot_history(); added arguments "show" and "ylabel", and dropped "suppress"
DOCUMENTATION
- Added section on "Exact Solution of the 1st Order Reaction A <=> B" to the Reactions page
- Updated all the Reference Guides, and overhauled their graphic appearance (especially their Tables of Contents navigation)
- Improved layout and navigation in the Experiments page
|
|
Nov. 4, 2023
Beta 29
|
CORE / NEW FEATURES
- Expanded support for enzymes
EXPERIMENTS
SUPPORTING LIBRARIES
- Split off the class
ThermoDynamics from "reactions.py" into its own file, "thermodynamics.py"
- Tighten check for identical reaction sides when instantiating class
Reaction
- Expanded the data now returned by
Reaction.extract_rxn_properties()
- Added object property "active_enzymes" to class
ChemCore; updated add_reaction() accordingly,
- The class
ChemData now also accepts a string as "name" argument, in case of a single chemical
- New method
ChemCore.all_chemicals()
- Tweaks to
ChemCore.get_all_names() and add_chemical_species()
- Renamed
ReactionDynamics.set_chem_conc() to set_single_conc()
VISUALIZATION
- Enhancement to
ReactionDynamics.plot_curves()
DOCUMENTATION
|
|
July 14, 2023
Beta 28.2
|
CORE
- Early support for macro-molecules : binding affinities (dissociation constants) and fractional occupancy based on ligand concentration
- Methods for macromolecule data struncture:
Macromolecules.add_macromolecules(), get_macromolecules(),
set_binding_site_affinity(), get_binding_site_affinity(), get_binding_sites(), get_binding_sites_and_ligands(),
get_ligand_name(), show_binding_affinities(), reset_macromolecule(), clear_macromolecules()
- Methods for macromolecule dynamic simulation:
ReactionDynamics.set_macromolecules(), set_occupancy(), get_occupancy(), update_occupancy(),
sigmoid(), logistic()
- New data class
ChemicalAffinity
- Added object variables
ReactionDynamics.macro_system_state and macro_system
- Exteded the method
describe_state(), to cover macromolecules
EXPERIMENTS
- New experiment, in folder "reactions_single_compartment/" :
macromolecules_1
(listing of all experiments)
- Updated all the other experiments, to reflect the change in the location of the
ChemData library
MAIN INFRASTRUCTURE
- New method
ReactionDynamics.get_chem_conc()
- Renamed constructor argument, and object variable, from "reaction_data" to "chem_data", in class
ReactionDynamics
SUPPORTING LIBRARIES
- Renamed "reactions/reaction_data.py" to "chemicals/chem_data.py"
- Dropped argument "n_species" from instantiation of class
ChemData
- Split off from the class
ChemData the new classes ChemCore, Diffusion, AllReactions, Macromolecules
- Diffusion data is now internally stored differently – and separately from other molecular data
- Dropped the object property
ChemData.n_species and the method _internal_reactions_data()
- New method
MovieTabular.clear_dataframe()
- Updated version of the
NeoAccess library to 4.3.1
VISUALIZATION
- New argument "range_x" to
ReactionDynamics.plot_curves(); tweaks to default colors
DOCUMENTATION
BUG FIX
- Bugfix in method
ReactionDynamics.set_conc()
EXAMPLE. From experiment
macromolecules_1:

|
|
June 4, 2023
Beta 27
|
CORE
- Support for enzymes, including data structure, detection, reporting, and efficient computing
EXPERIMENTS
- New experiments (all in folder "reactions_single_compartment/") :
enzyme_1 and state_space_1
MAIN INFRASTRUCTURE
- Changes to the class
ReactionDynamics, for increased modularity, in the context of the
new class Reaction
SUPPORTING LIBRARIES
- Split the class
ReactionData into ChemData and Reaction,
and more pytests
- Renamed some arguments in
ChemData.add_reaction()
- Tweaks to
ChemData.multiple_reactions_describe() and Reaction.describe()
VISUALIZATION
- New argument, "title_prefix", in method
ReactionDynamics.plot_curves()
BUG FIX
- Small bugfix in method
ReactionDynamics.single_compartment_react()
|
|
May 31, 2023
Beta 26.3
|
CORE
- Substantial refinement, polishing and expansion of the automated, adaptive variable time steps introduced in the previous release
- Eliminated the deprecated earlier approach of "dynamic substeps" (first introduced in Beta 18)
EXPERIMENTS
- Updated, verified - and revised as needed - all of the experiments
- Moved 4 single-compartment reaction experiments to a new "archived" subfolder
- Renamed experiment
substeps_2 to variable_steps_2
MAIN INFRASTRUCTURE
- The automated, adaptive step-size decisions are now guided by multiple norms (measures)
- Added a new norm based on the max relative concentration changes
- Major code reorganization and cleanup in the
ReactionDynamics class.
- Streamlined and expanded the diagnostic data, and its methods
- Simplifications to the
set_path.py script, and to all the notebooks that use it
- Reorganization of class
ReactionData
SUPPORTING LIBRARIES
- Early introduction of the Neo4j graph database, by means of the
NeoAccess library
from the sister open-source project BrainAnnex.org
- Added several new lookup options to
MovieTabular.get_dataframe()
- Upgraded
jupyterlab version to 3.4.8
- Changes to environment.yml, to install the needed Neo4j libraries
BUG FIX
- Bugfix in
ReactionDynamics.reaction_step_common()
- Bugfix in timestep change in case of "soft aborts" (automated re-try)
|
|
Mar. 6, 2023
Beta 25
|
CORE
- Introduced the option of automated, adaptive variable time steps, including supporting methods
such as
step_determiner_1() (implemented in class ReactionDynamics)
- Deprecated the earlier approach of "dynamic substeps" (introduced in Beta 18); they will be eliminated in the next beta release
EXPERIMENTS
- New experiments (all in folder "reactions_single_compartment/") :
up_regulate_3, variable_steps_1, substeps_1, substeps_2
- Small additions to experiment
negative_concentrations_1
MAIN INFRASTRUCTURE
- More diagnostics in case of steps aborted because leading to overly large concentration changes, or to negative concentrations
- New methods
get_diagnostic_rxn_data(), get_diagnostic_conc_data(), get_diagnostic_delta_data(), _delta_conc_dict()
- Added `silent` argument to
ReactionDynamics.explain_time_advance(); step sizes are now returned as well
- The values produced by the function
diagnostic_delta_conc_data are now saved during the run, rather than re-computed
- New object attribute
ReactionDynamics.diagnostic_delta_conc_data
- Renamed object attribute
ReactionDynamics.diagnostic_data to diagnostic_rxn_data
- Arguments `variable_steps` and `n_steps` no longer both allowed in
ReactionDynamics.single_compartment_react()
- Additional pytests
VISUALIZATION
- New method
plot_step_sizes()
- Tweaks to
plot_curves(), including new `show_intervals` argument
DOCUMENTATION
BUG FIX
- Small bugfix to
single_compartment_react()
EXAMPLE. From experiment
variable_steps_1:
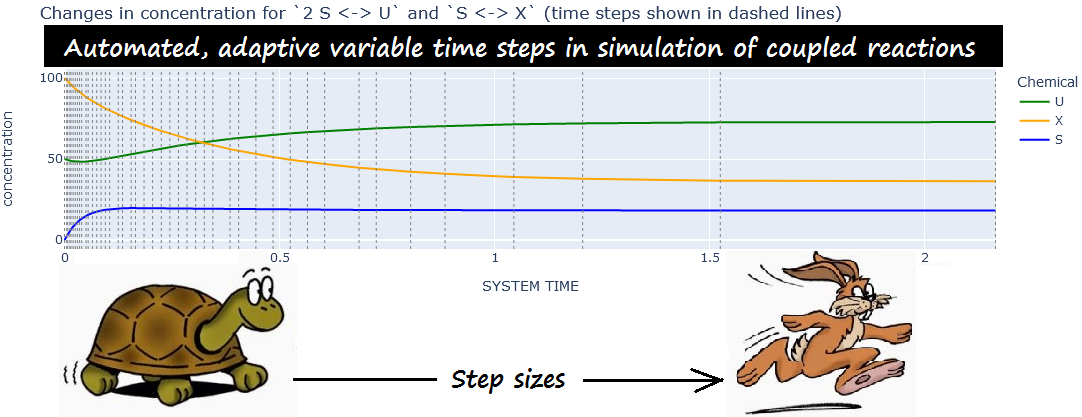
|
|
Feb. 11, 2023
Beta 24
|
CORE
- A more thorough check for - and automatic remediation of - concentrations values dipping into negative values
EXPERIMENTS
- New experiments
reactions_single_compartment/up_regulate_2 , large_time_steps_2 and negative_concentrations_1
MAIN INFRASTRUCTURE
- Expanded output in
ReactionDynamics.explain_time_advance()
- Change in default values for arg "snapshot" in
ReactionDynamics.set_conc() and set_chem_conc()
- New experimental method
_reaction_elemental_step_SINGLE_REACTION()
- Removed method
explain_time_advance_OLD()
SUPPORTING LIBRARIES
- Dropped support for (confusing!) chemical indices in
ReactionData.add_reaction()
DOCUMENTATION
- Added new section about "Numerical Instability" on the reactions page
- Updated section on "Adaptive Variable Time Sub-steps" on that same page
BUG FIX
- Bugfix in
ReactionDynamics._explain_time_advance_helper()
EXAMPLE. From experiment
negative_concentrations_1:
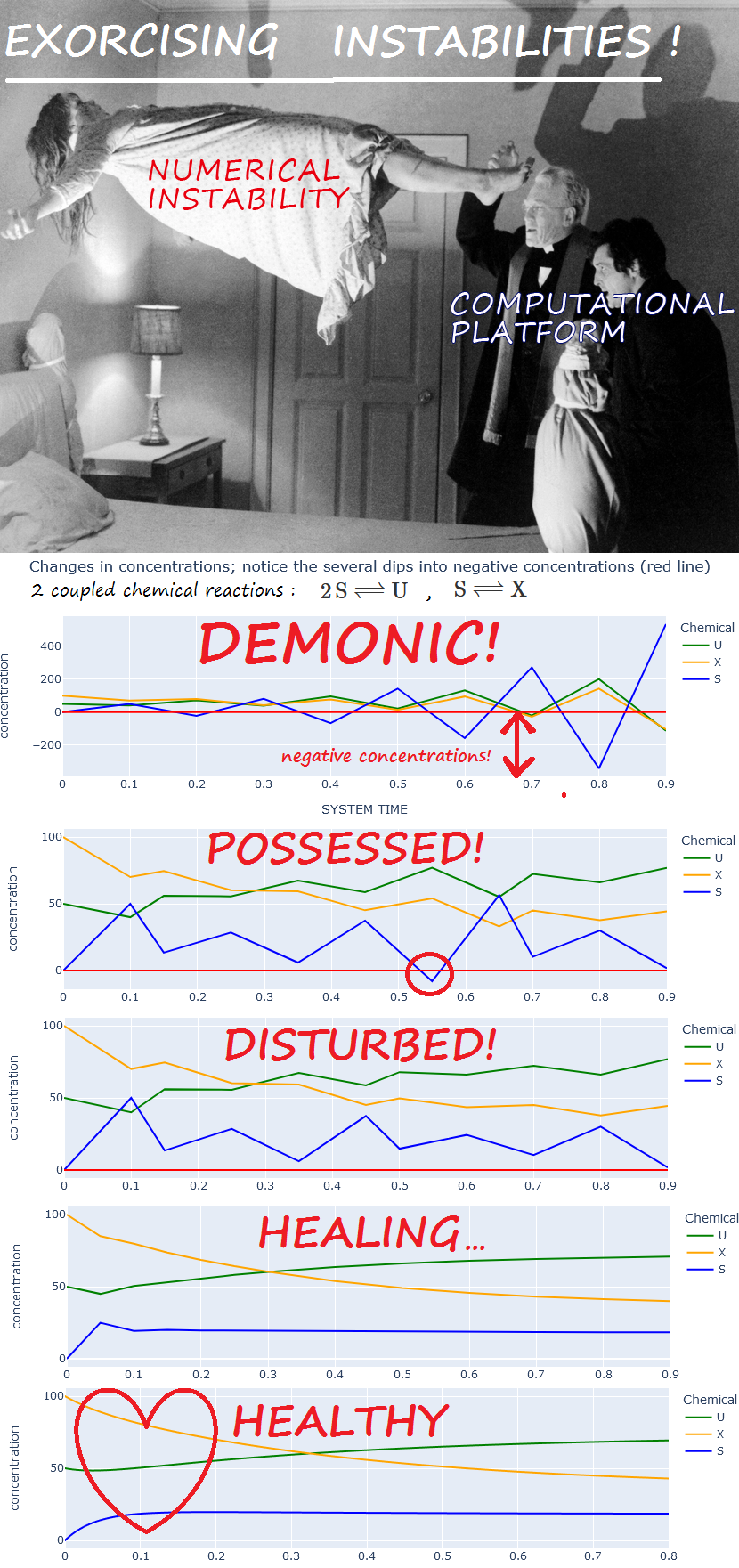
|
|
Feb. 5, 2023
Beta 23
|
CORE
- Major change in folder structure: all the core code and libraries
(exclusive of experiments and pytests) moved under top-level folder
src
EXPERIMENTS
- New experiment
reactions_single_compartment/impossible_1
- Updated several experiments in
reaction_single_compartment to utilize
newer functions such as plot_curves() and get_diagnostic_data(),
and to change argument name in single_compartment_react() from "fast_threshold" to "rel_fast_threshold"
MAIN INFRASTRUCTURE
- In
ReactionDynamics.single_compartment_react(), reactions get
reset to "fast" whenever the threshold (of what makes a reaction be regarded as fast) is lowered
- New method
set_rxn_speed_all_fast()
- Added new argument to
get_diagnostic_data() and to get_diagnostic_data()
- Tweak to
stoichiometry_checker_entire_run()
- In several methods in class
ReactionDynamics,
renamed argument "fast_threshold" to "rel_fast_threshold", and "dynamic_steps" to "dynamic_substeps"
- Renamed
reaction_step_NEW() to _reaction_elemental_step(),
and advance_variable_time_steps() to _advance_variable_substeps()
- Correction in status returned from
single_compartment_react()
- Added edge cases and pytests to
explain_time_advance()
SUPPORTING LIBRARIES
- Additional argument "Delta_G" to
ReactionData.add_reaction()
VISUALIZATION
- New argument to
ReactionDynamics.plot_curves()
EXAMPLE. From experiment
impossible_1:
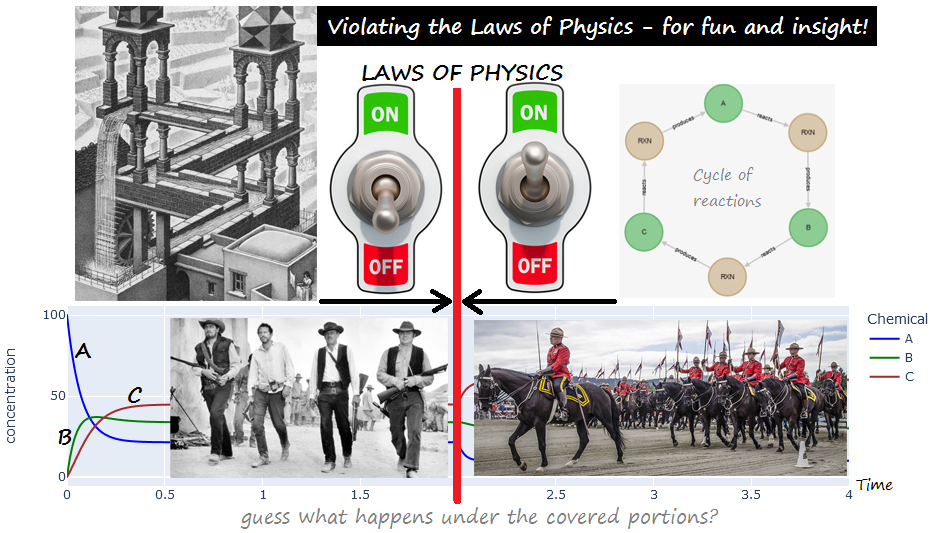
|
|
Jan. 30, 2023
Beta 22
|
CORE
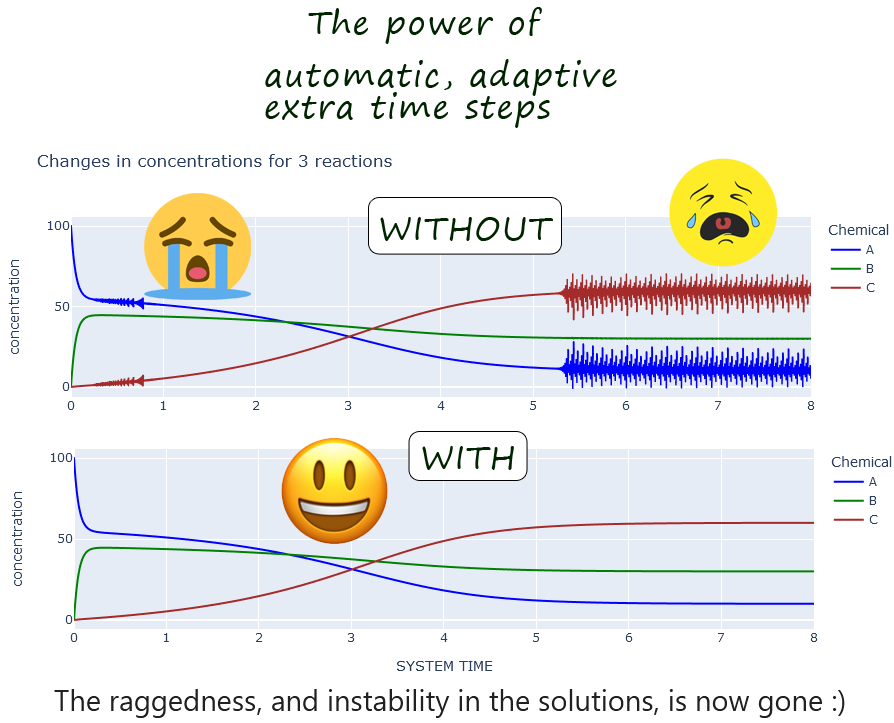
- Automatic throttling down of time-step size, as needed, to catch and immediately remedy errors from negative concentrations
arising from excessive time steps (including pytests)
EXPERIMENTS
- New experiments:
reactions_single_compartment/up_regulate_1
and reactions_single_compartment/large_time_steps_1
- Corrections & enhancements to experiments
reactions_single_compartment/cycles_1, down_regulate_1
- Small corrections/tweaks to experiments
reactions_single_compartment/down_regulate_2 , 1D/reaction/down_regulation_1 , 1D/reaction/up_regulation_1
MAIN INFRASTRUCTURE
- Improvements, including better precision, to
ReactionDynamics.curve_intersection()
- Added features to
ReactionDynamics.get_history()
- Improved status reporting of
single_compartment_react()
- New function
save_diagnostic_data()
- Consolidated the 2 overlapping functions
reaction_step_FIXED_RESOLUTION() and
reaction_step_VARIABLE_RESOLUTION() into reaction_step_NEW()
- Introduced custom Exception
ExcessiveTimeStep in class ReactionDynamics;
used it in reaction_step_orchestrator(), to catch and automatically remedy such errors
SUPPORTING LIBRARIES
- New functions
Numerical.segment_intersect(), line_intersect(), curve_intersect_interpolate()
BUG FIX
- Bugfix, and simplifications, in
ReactionDynamics.explain_time_advance()
EXAMPLE. From experiment
large_time_steps_1:
|
|
Jan. 24, 2023
Beta 21
|
CORE
- Major change in the criterion of tagging a reaction as "fast", implemented in the new function
criterion_fast_reaction().
This new approach has led to much-better results.
EXPERIMENTS
- New experiments:
cycles_1 and reactions_single_compartment/down_regulate_2
- Tweaks to experiments
react_2, react_3, and react_4
MAIN INFRASTRUCTURE
- New functions
ReactionDynamics.curve_intersection() and set_chem_conc()
- Expanded reaction diagnostic data, and new function
explain_time_advance() to utilize it
- Reaction speeds are reset whenever concentrations are altered
- New argument "abs_fast_threshold" to
single_compartment_react()
- Added "t_start" and "t_end" arguments to
get_history()
- Added argument "stop_time" to
single_compartment_react()
- Added argument "species_name" to
set_chem_conc()
- Now allowing dict argument to
set_conc()
- Change to return value of
is_in_equilibrium()
- Tweaks to
examine_increment_array(), get_conc_dict() and reaction_in_equilibrium()
SUPPORTING LIBRARIES
- New functions
Numerical.deep_flatten() and compare_results()
- Renamed
ReactionData.assert_valid_index() to assert_valid_species_index()
- Tweaks to
assert_valid_rxn_index() and to get_reaction()
- Added argument "tail" to
MovieTabular.get()
VISUALIZATION
- New function
ReactionDynamics.plot_curves()
DOCUMENTATION
BUG FIX
- Bugfix and pytests for
ReactionData.get_chemicals_in_reaction()
EXAMPLE. From experiment cycles_1:
|
|
Jan. 16, 2023
Beta 20
|
CORE
- Major refinement in the handling of the variable substeps for the time resolution of the reactions
EXPERIMENTS
- New experiment
reactions_single_compartment/down_regulate_1
- Renamed experiment
react_5 to cascade_1
MAIN INFRASTRUCTURE
- New function
ReactionDynamics.advance_variable_time_steps()
- Enhancements to function
reaction_step_VARIABLE_RESOLUTION()
- Replaced function
examine_increment() with a new approach, implemented in
examine_increment_array()
- Streamlined the function
reaction_step_orchestrator()
- The "tolerance" argument of
is_in_equilibrium() changed from absolute to relative
SUPPORTING LIBRARIES
- New function
ReactionData.get_chemicals_in_reaction()
- Tweaks to
describe_reactions() and its helper functions
- Removed the deprecated class
Movie
DOCUMENTATION
- New section and diagram on "Adaptive variable time substeps" added to the Experiments page
BUG FIX
- Bugfix to
ReactionData.add_chemical() and to ReactionDynamics._explain_reactions_helper()
EXAMPLE. From experiment
down_regulate_1:
|
|
Jan. 9, 2023
Beta 19
|
CORE
- Deeper scrutiny (and optional debug mode) of time substeps for reactions
- Extensive additional diagnostic information for adaptive time substeps
EXPERIMENTS
- New experiments:
reactions_single_compartment/react_3, react_4, react_5
- Expanded experiment
1D/reaction/reaction1
- Small enhancements to experiments
1D/reaction/reaction_4
- Tweak to experiment
1D/reaction/reaction_7
- Update most experiments of the
1D/reaction_reaction_x types, to save more results in the histories and plots
- Small additions, utilizing
stoichiometry_checker(), to experiments 1D/reaction/reaction_6 and 1D/reaction/reaction_7
MAIN INFRASTRUCTURE
- New methods added to the
ReactionDynamics class:
stoichiometry_checker(), clear_reactions(),
diagnose_variable_time_steps(), get_historical_concentrations(),
stoichiometry_checker_from_deltas(), stoichiometry_checker_entire_run(), delta_names(),
explain_reactions(), examine_run()
- Efficiency improvements to
single_reaction_step() : more detailed optional debugging output,
and split into 2 versions, for fixed time resolutions and for time substeps
- Change in argument name from "total_duration" to "reaction_duration" in
single_compartment_react(),
and from "delta_time" to "delta_time_full" in reaction_step_orchestrator()
- Split
examine_increment() into 2 functions; adjust pytests accordingly
- Introduction of the
self.diagnostics attribute in the class ReactionDynamics; misc. enhancements to diagnostics options
- Generalized
is_in_equilibrium() to multiple reactions, and used it in all the "react_X" series of experiments
- Tweaks to
compute_all_reaction_deltas(), and needed adjustments to pytests and to calling code
- Changed the
ReactionDynamics.diagnostic_data attribute to be a Python dictionary, with an entry per reaction.
- Switched the class
ReactionDynamics to use a "verbose_list" attribute
in lieu of the earlier "debug_code" and "debug" attributes
- New pytests for adaptive time substeps in reactions
- A few more validations across various functions in the class
ReactionDynamics
- Renamed
BioSim1D.save_snapshot() to add_snapshot()
SUPPORTING LIBRARIES
- Tweaks to
ReactionData.describe_reactions()
- Additional validations in
add_reaction(), with pytests
- Deprecated and completely phased out the class
Movie
BUG FIX
- 2 bugfixes in class
ReactionDynamics
- Fixed bug in 2 pytests
EXAMPLE. From experiment
cascade_1:
|
|
Dec. 21, 2022
Beta 18
|
CORE
- Adaptive variable time substeps for reactions (implemented in class
ReactionDynamics)
- Substantially expanded and revised the class
ReactionDynamics, including data structure management, data snapshots,
display of thermodynamic quantities, and estimation of reaction speeds. Pytests
EXPERIMENTS
- New experiment
reactions_single_compartment/react_2, to demonstrate adaptive variable time substeps for reactions
- Expanded experiment
reactions_single_compartment/react_1
LIBRARIES
- 2 new classes:
MovieGeneral and MovieTabular, plus tweaks to MovieArray, with pytests
- Dropped the deprecated libraries
Reactions and Chemicals
DOCUMENTATION
|
|
Nov. 28, 2022
Beta 17
|
CORE
- Elevated "single-compartment reactions" to top-tier module, alongside simulations in 1D, 2D and 3D.
LIBRARIES
- Merged the classes
Reactions and Chemicals (now both deprecated.)
Their merger was split into 2 new classes: ReactionData (to managed the data structures for the chemicals and the reactions)
and ReactionDynamics (to simulate the dynamics of reactions)
Pytests for the new classes.
- Expanded the
ReactionData class to manage thermodynamic data (Enthalpy, Entropy, Gibbs free energy)
- New method
ReactionDynamics.single_compartment_react()
- New methods
ReactionData.missing_diffusion_rate() and set_diffusion_rate()
INFRASTRUCTURE
- Extensive changes throughout, to deal with the restructuring of the libraries described above
EXPERIMENTS
- Updated all experiments, as needed by the restructuring of the libraries described above
- New experiment directory "reactions_single_compartment"
- New experiment
reactions_single_compartment/react_1
DOCUMENTATION
|
|
Oct. 17, 2022
Beta 16
|
CORE
- Introduced diffusion in 2D. New BioSim2D methods:
diffuse(), diffuse_step(), diffuse_step_single_species(), convolution_5_point_stencil(), system_snapshot(), system_size(), describe_state(), lookup_species()
inject_conc_to_bin(), set_species_conc(), with pytests.
- New BioSim1D methods:
inject_gradient(), inject_bell_curve(), system_size(), with pytests
LIBRARIES
- New method
Numerical.expand_matrix_boundary(), with pytests
EXPERIMENTS
- New 2D experiment:
2D/diffusion/diffusion_1
- New 1D experiments:
1D/diffusion/diffusion_along_gradient_1, 1D/diffusion/diffusion_along_gradient_2
- Updated experiment
1D/diffusion/gradient_1
VISUALIZATION
- Visualization for 2D system states, using plotly
- New method
BioSim1D.visualize_system()
DOCUMENTATION
INFRASTRUCTURE
- Split off pytests for diffusion in 1D into a separate file:
test_biosim_1d_diffusion.py
|
|
Oct. 7, 2022
Beta 15
|
CORE
- New methods
BioSim1D.diffuse_step_single_species_5_1_stencils(),
and double_spacial_resolution_linear(), with pytests
BioSim1D.diffuse(), and related methods, now accept an "algorithm" argument
- Added argument "copy" to
BioSim1D.lookup_species()
- Generalized
BioSim1D.is_excessive() and max_time_step(), to allow for arbitrary delta_x
- Updated the class
BioSim2D to conform to changes recently made to BioSim1D:
both classes are now designed to be instantiated
- Re-organization of the methods of
BioSim1D class; moved compare_states() to class Numerical
- Ditched
self.verbose from BioSim1D
LIBRARIES
- New class
Numerical,
with methods gradient_order4_1d(), gradient_order4(), and compare_vectors(), with pytests
- New class
MovieArray with 2 methods and pytests
EXPERIMENTS
- New trio of experiments:
1D/diffusion/validate_diffusion_1, _2, and _3
- Expanded the experiment
1D/diffusion/overly_large_single_timestep
- Updated the experiment
1D/diffusion/spacial_resolution_1
- Brought the experiment
2D/reaction/reaction_1 up-to-date with recent changes, and fixed a bug
INFRASTRUCTURE
- More pytests for
BioSim1D.diffuse_step_single_species() and for diffuse()
DOCUMENTATION
- Expanded the section on Diffusion in 2-D
- Added 3 diagrams to the site's home page
|
|
Sep. 14, 2022
Beta 14.1
|
INFRASTRUCTURE
- Incorporated delta_x (grid spacing) into 1D diffusion computations
- Converted class
BioSim1D from static to instantiated
(refactored all notebooks and pytests)
- Add method
BioSim1D.frequency_analysis(), with pytests, to provide frequency-component analysis with Discrete Fourier Transforms
- Add SciPy to the project requirements
- New method
BioSim1D.inject_sine_conc(), with pytests
- New methods
BioSim1D.save_system(), restore_system(), compare_states()
- The various
BioSim1D methods that change spatial resolution, now update the system state
- Added new argument to
BioSim1D.set_species_conc() ; slight tweaks to other methods that set concentrations
- Renamed method
Movie.append() to store()
EXPERIMENTS
- New experiments
1D/diffusion/low_pass_1 and 1D/diffusion/low_pass_2
- New experiment
1D/diffusion/spacial_resolution_1
- Added plotly heatmaps to experiment
1D/diffusion/gradient_1
DOCUMENTATION
BUG FIX
- Bugfix in
BioSim1D.inject_conc_to_bin()
|
|
Aug. 31, 2022
Beta 13
|
INFRASTRUCTURE
- Extended support for membranes in 1D, including expanded data structure in class
BioSim1D,
and expansion of several methods for data management, visualization and reactions:
set_membranes(), show_membranes(), describe_state(), set_bin_conc(), set_uniform_concentration(), lookup_species(), bin_concentration(),
react(), reaction_step()
New methods: uses_membranes() and bins_with_membranes()
- Implemented
system_length and mapping bin numbers to x-coordinats in class BioSim1D,
incl. class variable global_Dx , and new methods set_dimensions() and x_coord()
- New utility methods:
assert_valid_bin_number(), reset_system()
- Changed name of argument in
BioSim1D.set_bin_conc() and inject_conc_to_bin() from "bin" to "bin_address";
refactored all notebooks
- Changed default of "zero_clip" argument in
inject_conc_to_bin()
- More pytests, and general reorganization of them
EXPERIMENTS
- New experiment
1D/reaction/membrane_1
- Added plotly heatmap to experiment
1D/diffusion/diffusion_1
- Simplified experiment
1D/diffusion/reach_equilibrium_1
DOCUMENTATION
|
|
Aug. 22, 2022
Beta 12
|
INFRASTRUCTURE
- Implementation of reactions in 2D.
Added several methods to class BioSim2D, and pytests, for management of data structure, and for reactions
- Major re-organization, to take out of
BioSim1D any reaction-related methods also applicable to higher dimensions;
relocated several to Reactions
- Added "(state) snapshots" feature to
BioSim1D.react()
Reactions class: new methods is_in_equilibrium() and define_series()
- Early implementation of methods for the data structure to be used to model membranes
- Parallel tweaks to the constructors of
BioSim1D and BioSim2D
- Renamed argument `time_duration` of
BioSim1D.diffuse() to `total_duration`
- New method
Chemicals.assert_valid_index() ; simplifications to BioSim1D using it
Tweak to Chemicals.get_index(); added extra validation and pytests for the class
EXPERIMENTS
- First 2-D experiment:
2D/reaction/reaction_1
- New experiments:
1D/reaction_diffusion/state_space_1, 1D/reaction/up_regulation_1, 1D/reaction/down_regulation_1
- Renamed the experiment folders from
life_1D to 1D, to avoid confusion with the location of
the simulation libraries. Likewise for life_2D
- Streamlined most experiments by using the new method
Reactions.is_in_equilibrium()
- Simplifications and additional visualizations to experiment
1D/diffusion/diffusion_1
VISUALIZATION
- Expanded the class
GraphicLog to allow use of graphic libraries stored remotely,
to avoid a JupyterLab bug preventing the correct display of HTML log files with links to locally-stored files (typically .CSS and .JS),
when using the "Open in New Browser Tab" feature in JupyterLab.
Changed all experiments to use remote linked files in the HTML log files they create
- Broader use of
plotly (only plotly EXPRESS had been used thus far)
|
|
Aug. 13, 2022
Beta 11.3
|
INFRASTRUCTURE
- Introduced wide support for SYSTEM TIME throughout
BioSim1D
- New methods to facilitate creation and use of snapshots of the state of the systems,
incl.
system_snapshot(), bin_snapshot(), show_system_snapshot(),
save_snapshot() and get_history()
- Added "tabular" option to
Movie class. Incorporated Movie object in BioSim1D class
- In class
Reactions, added argument to describe_reactions()
- Added new argument to
BioSim1D.set_uniform_concentration()
- Started using Jupytext: added it to
requirements.txt , and added config file jupytext.toml .
All notebooks are now linked to .py files by the same name.
This is useful to refactor notebooks upon changes in the underlying code libraries
(discussion)
EXPERIMENTS
- New experiment:
diffusion/gradient_1
- Added extra visualization with
plotly to most existing experiments
- Revised, added comments, and simplified most existing experiments, in particular to take advantage of the new support for SYSTEM TIME
VISUALIZATION
- Added the visualization libraries
plotly and matplotlib to the standard setup,
and provided support for plotly.
- Added support for arbitrary color mapping to
vue_curves_4, incl. new method pale_version()
- Added argument `color_mapping` to
BioSim1D.line_plot()
- Changes to
vue_cytoscape_1 and HtmlLog.export_plot_Vue() to allow multiple network plots on same page
DEPLOYMENT
- For compatibility with optional deployment to mybinder.org, added
file
environment.yml, to specify Python version 3.8 and other requirements
(details)
DOCUMENTATION
|
|
July 13, 2022
Beta 10
|
VISUALIZATION
- New Vue/D3 component
vue_cytoscape_1 (with tests), for visualizing networks ("directed graphs"), with re-positionable and clickable nodes. Based on the open-source cytoscape.js library.
EXPERIMENTS
INFRASTRUCTURE
-
Changed HtmlLog.config() and GraphicLog.config(), to accept multiple JavaScript files, to incorporate links to the cytoscape.js library.
-
Enhancements, incl. new methods, to class Reactions; in particular, to generate visualization data
-
Added methods number_of_chemicals() and get_diffusion_rate() to class Chemicals.
-
Changed file naming conventions in experiments/visualization/logs_with_Vue_component_tests
-
Added and updated pytests
DOCUMENTATION
|
|
July 6, 2022
Beta 9.1
|
VISUALIZATION
EXPERIMENTS
INFRASTRUCTURE
- Switched to python 3.8
New method BioSim1D.line_plot(), to provide support for the new graphic module vue_curves_4 Cleanup and various enhancement to class Chemicals, plus pytests for most of its methods Tweaks to BioSim1D.describe_state(), and corresponding adjustments to affected notebooks Tests on the use of the multiprocessing library (not yet included in main repository)
DOCUMENTATION
|
|
June 13, 2022
Beta 8.2
|
DEPLOYMENT
- Life123 can now be
TRIED LIVE at Binder, with 1 click!
(akin to Google's Colab: short-term runs on a hosted JupyterLab environment)
- Introduced several utilities, and made corresponding changes to all notebooks, to work on Binder.
In particular, forked and modified the notebook utility library ipynbname , and also changed all notebooks to utilize the new set_path.py utility
INFRASTRUCTURE
- New class
Movies , to organize snapshots of system data for combined visualization
DOCUMENTATION
- Documented the newly-available Binder hosting environment
|
|
June 4, 2022
Beta 7
|
EXPERIMENTS
- All experiments with heatmap plots now expanded to also show results as line charts (and interpolating curves) with the new graphic module
vue_curves_3
- Reorganization of 1D diffusion experiments
VISUALIZATION
- New Vue/D3 component
vue_curves_3 , with tests
- Simplified the combined use of
HtmlLog and Vue components with new class GraphicLog
- In class
BioSim1D, the method single_species_heatmap() now makes use of the new GraphicLog class. Fixes to allow coexistence of multiple plots. Added method single_species_line_plot()
- Reorganization of visualization files, and streamlining of naming convention
- New methods for horizontal axes added to library SVGhelper (now in version 1.2)
INFRASTRUCTURE
- New utility file
get_notebook_info.py , to extract notebook names at runtime
- Tweaks to error messages in class
HtmlLog
- Prepared the website for future interface with a BrainAnnex-based content management system
DOCUMENTATION
- Greatly expanded website sections on "Visualization" and "Experiments", including summaries of all experiments
- Expanded README
|
|
May 13, 2022
Beta 6
|
EXPERIMENTS
- Revised all applicable notebooks to make use of the improved graphic component
heatmap-11 (with range slider and value tooltips)
- Simplified all applicable notebooks by making use of the recently-added method
BioSim1D.single_species_heatmap()
VISUALIZATION
- New Vue/D3 components:
heatmap-10, heatmap-11 and curves-1
- Expansion and enhancements to library SVGhelper (now in version 1.1)
- Reorganization of tests for visualization components
LIBRARIES
- Simplification of the data handover from Python to Vue.js in HtmlLog (to log graphical elements)
DOCUMENTATION
|
|
Apr. 30, 2022
Beta 5
|
INFRASTRUCTURE
- Converted all the remaining Python files for the experiments into Jupyter Notebooks
- Re-oriented from the classic Jupyter Notebooks to JupyterLab. New batch file:
quick.bat to start JupyterLab
- New utility/testing notebooks: "diagnostics" and "html_display_test"
BUGFIXES
- Bugfixes to the early experiments
- Restored the linear scale for heatmaps in the Vue component
heatmap9.js
LIBRARIES
- Enhancements to HtmlLog; in particular, to operate with JupyterLab notebooks
CORE
- Tweaks to the BioSim1D and Reactions classes
|
|
Apr. 17, 2022
Beta 4
|
- Much-expanded support and documentation for Jupyter Notebooks (used for the experiments); in particular, a major new "How to Run" section in the README, a
quickstart.bat file, and a diagnostic script.
- Continued conversion of the Python files for the experiments into Jupyter Notebooks
|
|
Apr. 8, 2022
Beta 3
|
- New methods to increase/decrease spatial resolution (1D)
-
New methods replace_system(), in 1D, and initialize_system(), in 3D
-
Introduction of Jupyter Notebooks for the experiments
-
General re-organization
|
|
Mar. 15, 2022
Beta 2
|
- Introduced combined reaction-diffusion in 1D
- Both the diffusion and reaction steps are now non-destructive on the system data, and buffer the "Delta concentrations"
- Early development for a new class dealing with "heuristics" for deciding accuracy parameters
|
|
Mar. 14, 2022
Beta 1
|
Initial public release
|
Alpha versions:
|
|
Dec. 2021 -
Mar. 2022
Alpha (late)
|
- Early draft of data structure for systems in 1-, 2- and 3-D
- Diffusion and reactions in 1-D, with testing and heatmap visualization
- A module for HTML logs, integrated with Vue/D3 visualization
|
|
Spring 2020
Alpha (early)
|
Early prototypes
|










